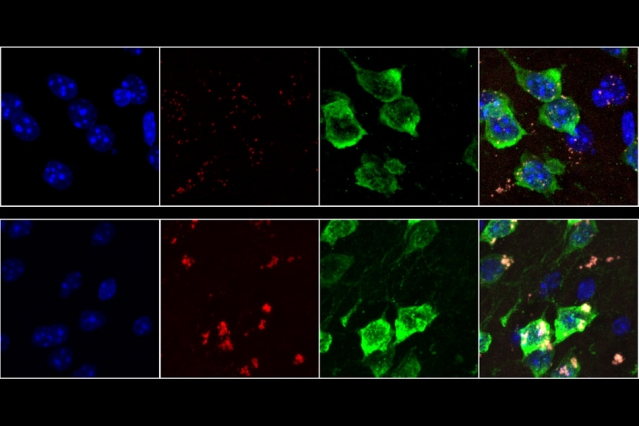
Neurons from 19-month-old mice (bottom row) show much higher rates of oxidative stress than neurons from 9-week-old mice (top row). Lipofuscin, a molecule associated with aging-related oxidative stress, appears red. Image: Hyeseung Lee
Source: [Anne Trafton | MIT News Office, November 27, 2018]
Snippets of RNA that accumulate in brain cells could interfere with normal function.
As we age, neurons in our brains can become damaged by free radicals. MIT biologists have now discovered that this type of damage, known as oxidative stress, produces an unusual pileup of short snippets of RNA in some neurons.
This RNA buildup, which the researchers believe may be a marker of neurodegenerative diseases, can reduce protein production. The researchers observed this phenomenon in both mouse and human brains, especially in a part of the brain called the striatum — a site involved in diseases such as Parkinson’s and Huntington’s.
“The brain is very metabolically active, and over time, that causes oxidative damage, but it affects some neurons more than others,” says Christopher Burge, an MIT professor of biology. “This phenomenon appears to be a previously unrecognized consequence of oxidative stress, which impacts hundreds of genes and may influence translation and RNA regulation globally.”
Burge and Myriam Heiman, the Latham Family Career Development Associate Professor of Brain and Cognitive Sciences, are the senior authors of the paper, which appears in the Nov. 27 issue of Cell Reports. Peter Sudmant, a former MIT postdoc, is the lead author of the paper, and postdoc Hyeseung Lee and former postdoc Daniel Dominguez are also authors.
A mysterious finding
For this study, the researchers used a technique developed by Heiman that allows them to isolate and sequence messenger RNA from specific types of cells. Messenger RNA carries protein-building instructions to cell organelles called ribosomes, which read the mRNA and translate the instructions into proteins by stringing together amino acids in the correct sequence.
Heiman’s technique involves tagging ribosomes from a specific type of cells with green fluorescent protein, so that when a tissue sample is analyzed, researchers can use the fluorescent tag to isolate and sequence RNA from only those cells. This allows them to determine which proteins are being produced by different types of cells.
“This is particularly useful in the nervous system where you’ve got different types of neurons and glia closely intertwined together, if you want to isolate the mRNAs from one particular cell type,” Burge says.
In separate groups of mice, the researchers tagged ribosomes from either D1 or D2 spiny projection neurons, which make up 95 percent of the neurons found in the striatum. They labeled these cells in younger mice (6 weeks old) and 2-year-old mice, which are roughly equivalent to humans in their 70s or 80s.
The researchers had planned to look for gene expression differences between those two cell types, and to explore how they were affected by age. “These two types of neurons are implicated in several neurodegenerative diseases that are aging-related, so it is important to understand how normal aging changes their cellular and molecular properties,” Heiman says.
To the researchers’ surprise, a mysterious result emerged — in D1 neurons from aged mice (but not neurons from young mice or D2 neurons from aged mice), they found hundreds of genes that expressed only a short fragment of the original mRNA sequence. These snippets, known as 3’ untranslated regions (UTRs), were stuck to ribosomes, preventing the ribosomes from assembling normal proteins. “While these RNAs have been observed before, the magnitude and age-associated cell-type specificity was really unprecedented,” says Sudmant.
The 3’ UTR snippets appeared to originate from about 400 genes with a wide variety of functions. Meanwhile, many other genes were totally unaffected.
