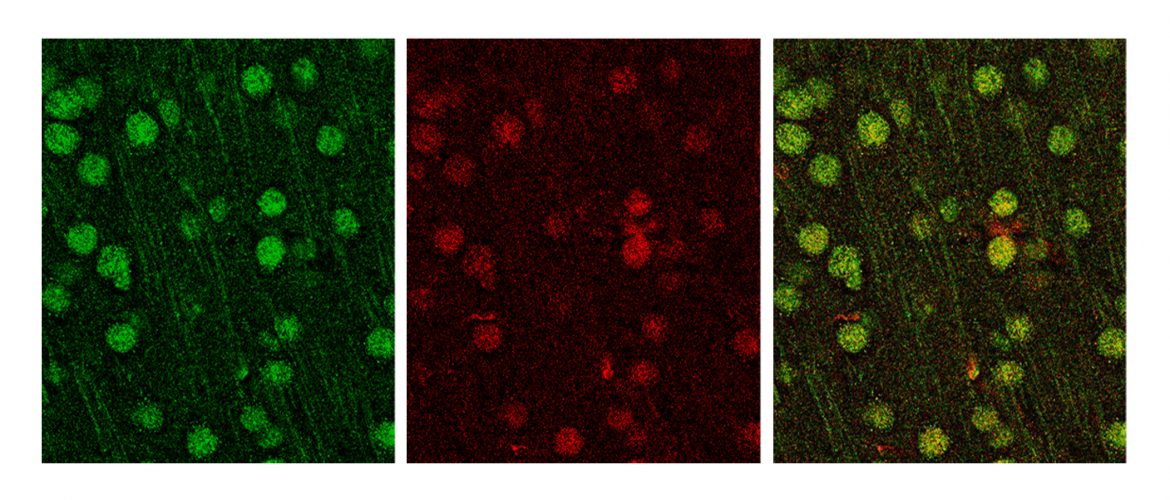
MVP immunostaining. MVP in green, neuronal marker SATB2 in red | Credit: Jacque Ip, Sur Laboratory, Picower Institute for Learning and Memory
Source: [Picower Institute for Learning and Memory, David Orenstein | March 20, 2018]
In a new study of one of the most common genetic causes of autism, neuroscientists at MIT’s Picower Institute for Learning and Memory have identified a specific molecular mechanism that appears to undermine the ability of neurons in affected mice to properly incorporate changes driven by experience. The findings published in the Journal of Neuroscience therefore suggest that a particular gene, MVP, is likely consequential in people with 16p11.2 deletion syndrome.
Accounting for up to 1 percent of autism cases, 16p11.2 deletion occurs in people who are missing a small region of DNA near the center of one copy of chromosome 16. For years, scientists have been working to determine exactly how the reduced presence of 29 protein-encoding genes leads to clinical symptoms of the syndrome such as autism-like behaviors, developmental delay and intellectual disability.
“This has been a major problem for the field,” said senior author Mriganka Sur, Newton Professor of Neuroscience in the Picower Institute and director of the Simons Center for the Social Brain at MIT. “People have looked at the entire region in mice. Our strategy was different. We thought, can we make a hypothesis about a critical gene that should play an important role?”
The team led by postdoctoral associate Jacque Pak Kan Ip focused on MVP, which is known for encoding a protein that shuttles RNAs and proteins from the nucleus to the rest of cells – a generally vital function. It is particularly important in the immune system and is known to regulate other genes that are as well. Though MVP is also known to be among the 29 affected in people with 16p11.2 deletion syndrome, its specific function in neurons has barely been investigated. To change that, the researchers devised a series of tests of MVP in a well-understood region in the visual cortex, where the brain processes sight from both eyes.
Keeping an eye open for change
Ip and co-authors employed the time-honored protocol of “monocular deprivation,” or temporarily shutting one eye for a week. Normally, responses in visual cortex neurons related to the closed eye become weaker, but responses related to the remaining open eye become stronger, as if to compensate for the change in ability. This neural circuit adjustment to experience is called “homeostatic plasticity.”
In the normal mice, closing an eye for a week had the expected effect. But in mice with one copy of MVPmissing, the researchers observed a telling difference. Responses related to the closed eye still weakened, but responses from the open eye did not get stronger. Homeostatic plasticity was disrupted.
“After seeing that MVP is responsible, we asked how MVP does it,” Ip said.
In those further tests, the team found that neurons in MVP-reduced mice experienced less electrical current across their excitatory synapse connections with other neurons, suggesting reduced functional excitatory synapses. They found other differences, too. MVP-reduced mice overexpressed the gene STAT1, an immune-system gene known to be regulated by MVP.
In 2014 Sur’s lab had found that mice without STAT1 have unusually strong open-eye responses after monocular deprivation. Now in the new study, with less MVP than usual and too much STAT1, mice experienced the opposite. Sure enough, when the researchers knocked down STAT1 as well as MVP, they were able to bring back a near-normal open-eye response to monocular deprivation, suggesting that loss of MVP disrupts homeostatic plasticity by allowing for an overabundance of STAT1.
Then Ip, Sur and co-authors dug even deeper. They found that in MVP-reduced mice, neurons weren’t producing the expected open-eye responses because they weren’t expressing a key receptor, the GluA1 AMPA receptor, on the surface of dendritic spines.
Even though the findings were made, by design, in the visual cortex, the authors said they expect the disruption of homeostatic plasticity to occur elsewhere as well.
“This gene’s presence is reduced everywhere in the brain,” Sur noted.